7 Comments
We were pleased to hear the feedback on our previous post on NIH-funded model-organism research. One question a number of you asked is: what’s happening with research involving mouse models? Thanks to additional work by colleagues in NIH’s Office of Portfolio Analysis (OPA) and Office of Extramural Research/Office of Research Information Systems, I’m excited to be able to share on the blog today how we approached this question.
We examined over 267,000 NIH grant applications submitted from FY2008 to FY2015, searching for the words “Mus musculus,” “mouse,” or “mice” in the descriptive text. Of these 267,000 applications, there were 190,329 applications (71%) that contained any of these keywords.
Figure 1 shows the number of new R01 awards for research projects using mouse models compared to all new R01 awards. The upward and downward patterns for research using mice reflect those of all research.
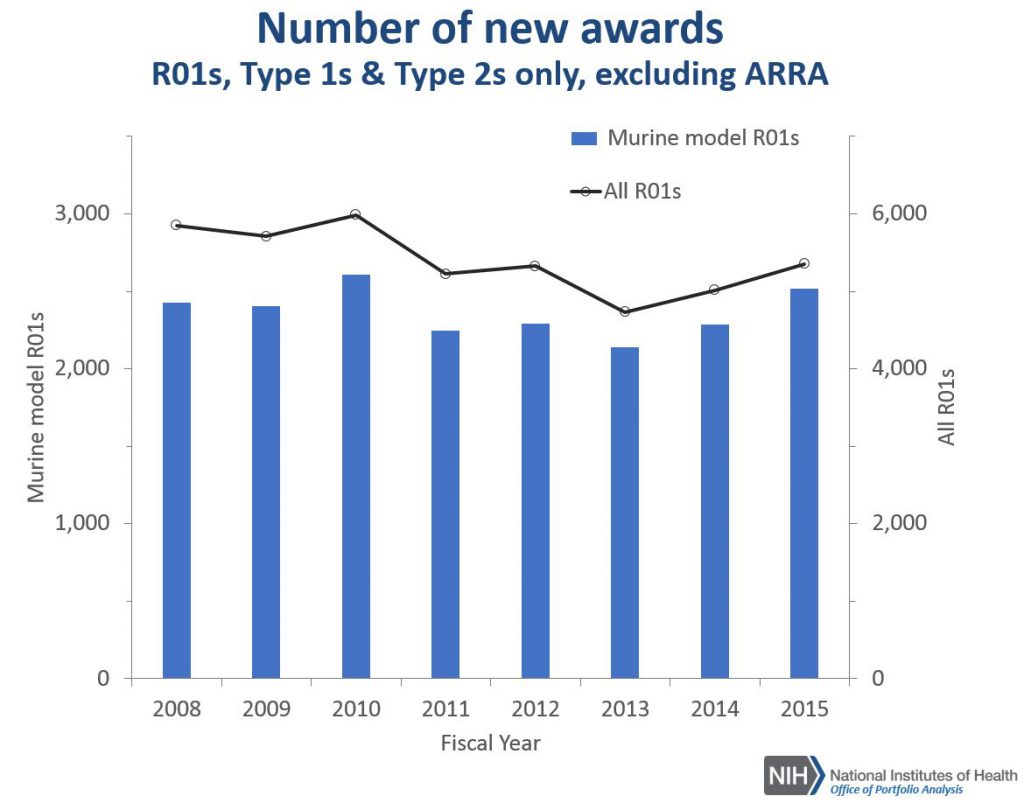
Figure 2 shows corresponding data for all research project grant (RPG) mechanisms. While the overall number of new RPG awards has been declining, the number of new projects using mouse models has increased.
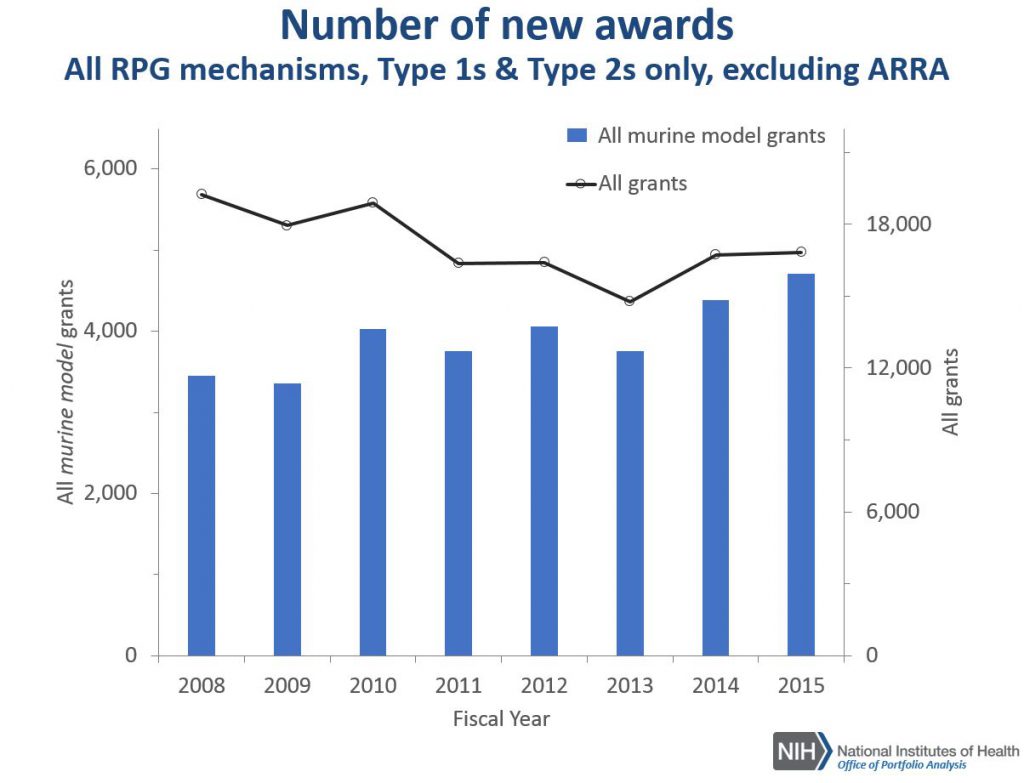
Figure 3 synthesizes the data from Figures 1 and 2 by showing what percentage of all R01/RPG awards involve mouse models. For both of these, we see increasing trends – each year, a greater proportion of new awards involve use of a mouse model.
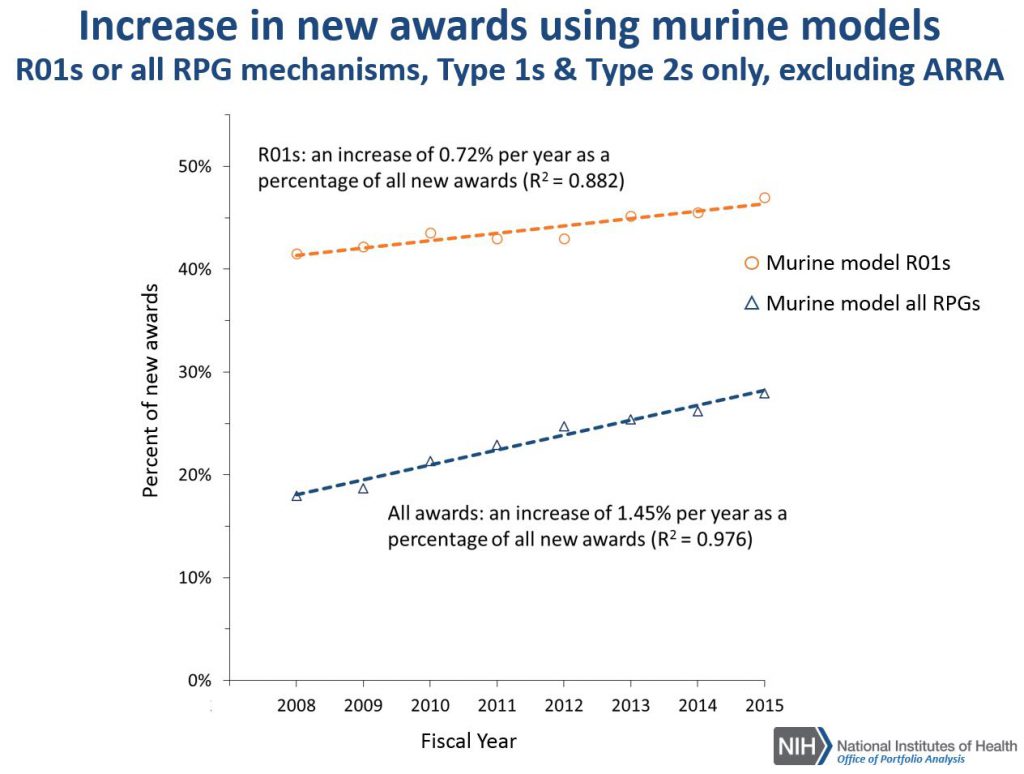
Figure 4 shows the award rates for competing R01 applications: there were no differences in award rates for R01s that employ a murine model compared to all R01s.
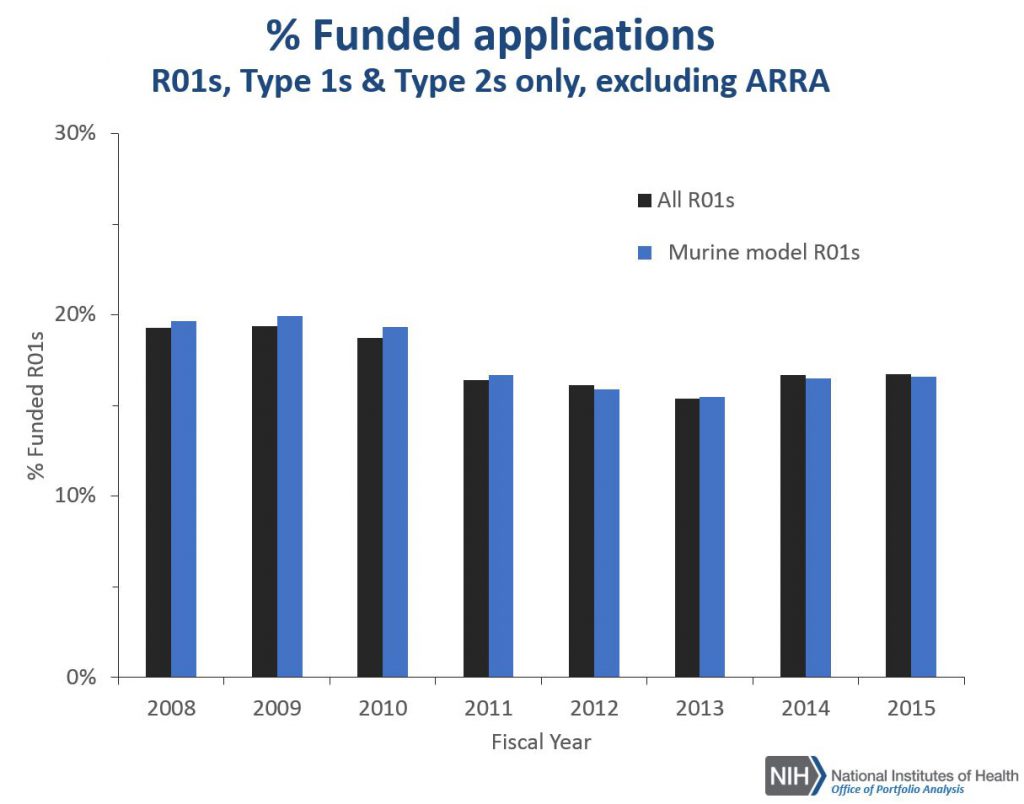
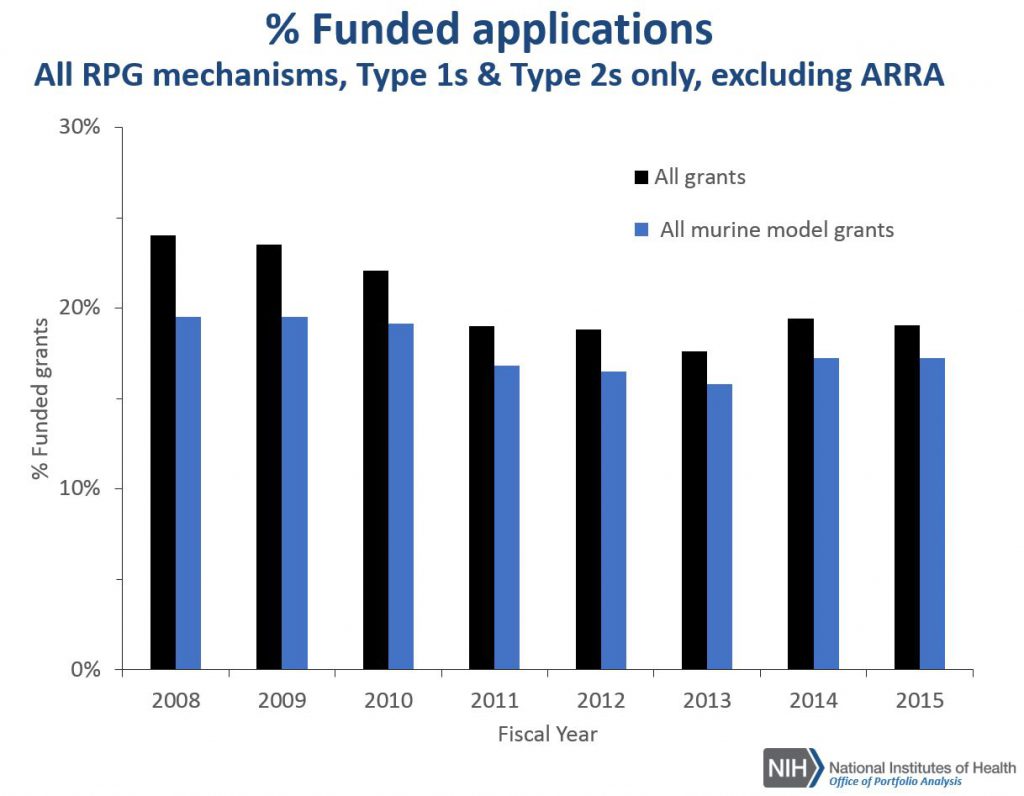
Figure 5 shows corresponding award rate data for all RPGs. RPG applications that employ mouse models have lower award rates compared to all RPGs, though the differences have narrowed over time. The lower award rate of applications for research that uses mouse models compared to all RPGs may at least in part explained by the higher percentage of those projects that are submitted as R01 applications; R01s have a lower award rate (regardless of the organism being studied) relative to all RPG activity codes.
In summary, it appears that if anything, research using mouse models is playing an increasing role in our extramural portfolio. These “model organism part 2” findings complement our previous findings on support for other model organisms. In a final “model organism part 3” blog, we will examine all-RPG support for multiple model organisms using analyses that rely on a different methodology than what we showed here in parts 1 and 2.
We are grateful to George Santangelo, Rick Ikeda, Judy Riggie, and our colleagues in the Office of Portfolio Analysis and the Office of Research Information Systems for their work on these and related analyses.



I have been concerned that much of the research on model organisms that NIH has supported has not been applicable to or meaningful for humans. I have been most interested in studies that use mice as models of human disease. Research with mice has been justified on the basis of their genetic homologies and physiological similarities to humans, but has not paid sufficient attention to the evolved differences between the two species. Mice and humans have very different phenotypes in the absence of disease, and so it is not surprising that they often differ in the pathways leading to disease phenotypes. I am attaching a reference to an article I recently published on the use of mice as models for studying human disease. I hope that the evolutionary considerations I discuss will inform future NIH support of model organism research.
Perlman, R.L. Mouse models of human disease: An evolutionary perspective. Evol Med Public Health. 2016 May 21;2016(1):170-6.
Thank you, Team Open Mike, for answering some of my questions from Part 1 in this Part 2 post. I hope you will answer all my other questions in Part 3 and provide a comprehensive comparison of the portfolio of organisms utilized by NIH biomedical research programs. This post confirms my hunch I mentioned in my previous comments – “mouse” gets a whopping 71% of ALL current NIH GRANTS, and comprising >45% of new R01 grants.
A more useful perspective is to compare this to the “WFFF” model organisms: (Worm, ~0.9%, Fly, ~2.2%, Fish, ~1.2%, Frog, ~0.6%), which together is just ~5%. The NIH funding portfolio is funding~10X more resources on the ‘mouse’ than on these “other” model organisms, which clearly only fit the category of “Basic/Fundamental” research, whereas ‘mouse’ studies can fit both categories of “Basic” and “Translational” research.
Regardless of how Basic and Translational research is measured for the mouse, it is dismaying to see such a paltry percentage of NIH funding support for the tried and true “Basic Research” model systems categorized here as “WFFF”. As a “well-rounded” scientist, I have actually worked with every one of these five organisms, each providing their unique value towards studying my favorite biological pathway. But I can attest that in no way has my mouse work yielded 10X more publications or insight to biology compared to my work in the other “WFFF” animals, not even close.
In my experience, I would argue NIH and humanity would continue to get more “bang for the buck” in terms of knowledge, productivity, and breakthroughs by equalizing the priorities and proportions of funding back towards “lower” organisms, from the “WFFF” animals down to our microbial communities. If the NIH truly stands by its pledge in the journal Science that “Basic science: Bedrock of progress”
then I would like to see the true action to this pledge by re-balancing more support to the model systems that only drive the “Basic/Fundamental” arms of the NIH research portfolio.
I wonder how accurate the 71% figure is….. searching for “mouse”, “mice” or “Mus musculus” in the descriptive text seems an imprecise way of getting the information. It would seem more pertinent to search the “Vertebrate Animals” section and/or the “Justification” subsection. The descriptive text may refer to prior work or studies of others used for justification. Also what percentage of application have a “Vertebrate Animals” section?Perhaps Team Open Mike can clarify the search criteria?
What percent of funded grants incorporate both “human and mouse” systems? I’m guessing it is more than “human and fly” or “human and worm”, which is potentially why there is a difference in funding here.
We’d really like to see the numbers for yeast!!! (Echoing an earlier comment). It is common wisdom that NIH study sections are now heavily biased against yeast despite the major breakthroughs in basic knowledge about eukaryotic cells. Let’s take a look at the numbers and see if conventional wisdom is right.
We greatly appreciate your suggestions regarding further analyses.
Yeast model organism support is covered in our “part 3” blog post.
I think there needs to be a discrimination between real mouse models of disease, such as genetically modified mice, and studies that use mice as fancy “petri dishes” to grow tumors. To me the latter are a different category of studies.
I echo the comment above that using the vertebrate animal section is a much better way than just searching for words in the grant.