5 Comments
Citations from scientific articles are more than lines on a page. They can, when reading between those lines, shed some light on the development of scientific thought and on the progress of biomedical technology. We’ve previously posted some examples in blogs here, here, and here. But to better see the light, we all would benefit from more comprehensive data and easier access to them.
My colleagues within the NIH Office of Portfolio Analysis sought to answer this call. Drs. Ian Hutchins and George Santangelo embarked on a hefty bibliometric endeavor over the past several years to curate biomedical citation data. They aggregated over 420 million citation links from sources like Medline, PubMed Central, Entrez, CrossRef, and other unrestricted, open-access datasets. With this information in hand, we can now take a better glimpse into relationships between basic and applied research, into how a researchers’ works are cited, and into ways to make large-scale analyses of citation metrics easier and free.
As described in their recent PLOS Biology essay, the resulting resource, called the NIH Open Citation Collection (OCC), is now freely available and ready for the biomedical and behavioral research communities to use. You can access, visualize, and bulk download OCC data as part of the NIH’s webtool called iCite (Figure 1). iCite allows users to access bibliometric tools, look at productivity of research, and see how often references are cited.
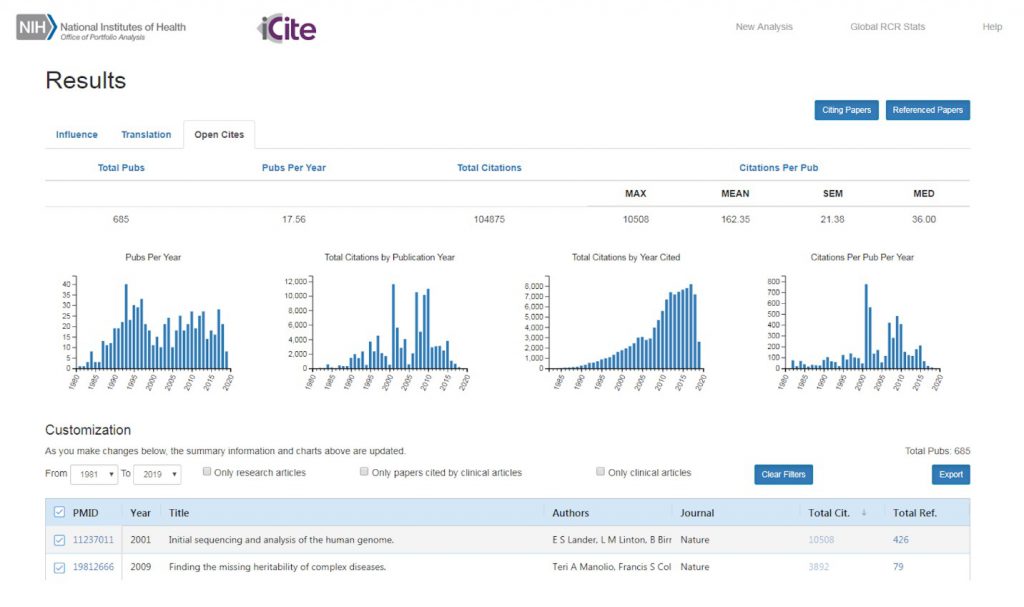
Figure 2 illustrates the new OCC web interface. Data from a group of publications are displayed on a summary table on the top. Various charts with visualizations lie beneath the summary table. They show publications over time (left), total citations per year by the publication year of the referenced article (center left) or the citing article (center right), and average citations per article in each publication year (right). These tables are customizable as publications are selected or deselected from the portfolio. You can also see information related to the article, such as links to the citing and referenced papers on PubMed, on the bottom of the screen.
The new OCC resource collection within iCite aims to reduce the costs of large-scale analyses of structured citation data, a recognized impediment for the bibliometrics field. OCC goes further still. It enhances the quality, robustness, and reproducibility of analyses using citation data. Moreover, it allows those interested to freely access structured data and share it with others. And, it also provides for transparency, which improves understanding of how knowledge flows and applied technologies develop.
Let’s use OCC to see that knowledge flow in action (Figure 3). Here the team assessed citation networks associated with the development of cancer immunotherapy. Each dot represents a scientific paper. The color represents whether the paper describes basic (green), translational (yellow), or clinical (red) science. The most influential clinical trials are shown in the large red dots in the center. These trials formed part of the evidence base FDA required for approval as a clinical treatment.
Information available in OCC will continue to grow. In addition to accumulating citations, the OCC will acquire data preprint servers and other materials currently not indexed in PubMed.
We invite you to take a look at and use the OCC. It will be exciting to see how the research community will use this new resource when conducting their own analyses. Data from these studies delving into citation dynamics may even provide additional insights that help all of us better understand how the scientific enterprise works and how we could make it even better.



Open citations collections (OCC)-NIH Grants USA snapshots were meticulously-presented with comprehensive citations’ amalgamation from prestigious scientific databases, Pubmed, Medline, etc. I gained expert critical research insights in first/lead authorships and ethical publication practices; the innovative strategies would be beneficial for global scientific integrity in authorships, projects, grants with patient-friendly eventual cost-effective public health research model!
Thank you for developing and sharing this resource. I am an independent researcher and am at a disadvantage for accessing bibliographic information that this resource offers. I will definitely explore this further.
This is very useful tool and very well thought off. However, as a scientist who publishes in the interface of chemistry and biology I see that 30-50% of citations to my papers don’t show up because most chemistry papers are published in journals not included in PubMed. I am quite sure something very similar would be happening to those working in the interface of biology and physics or information sciences. Perhaps including databases other than PubMed will alleviate this problem. Also unlike google scholar there is no option of aggregating articles from the same author when they are published under different names
Unfortunately, this tool is bias toward experimental scientists. Top highly scientists pursuing exclusively theoretical work are featured poorly Weighted RCR and percentile.
Very good progress is being made. We are pleased to see these promising results.
Thank you very much!