10 Comments

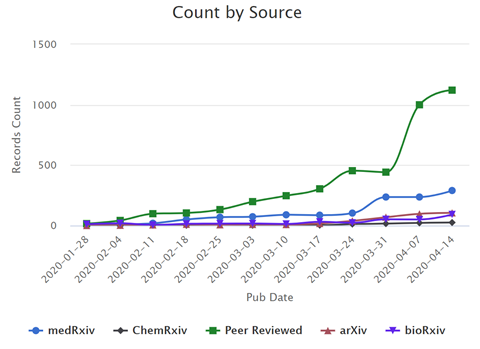
In the past few months, the scientific community has ramped up research in response to the SARS‑CoV‑2 pandemic; dozens of peer-reviewed articles and preprints on this topic are being added to the literature every day (Figure 1). This rapidly expanding effort has created challenges for scientists and the medical community who need to analyze thousands of scholarly articles for insights on the virus.
Recently, the National Library of Medicine at NIH joined the White House and key industry and university leaders to release the COVID-19 Research Dataset (CORD-19) and call on the AI community to develop text mining tools that help analyze and summarize the over 45,000 coronavirus articles. The CORD-19 dataset represents the most comprehensive, freely available library of machine readable coronavirus scholarly literature to date, with hundreds of AI tools and technologies already created.
Building on this effort, the NIH Office of Portfolio Analysis (OPA) has assembled a comprehensive listing of COVID‑19 publications and preprints that is freely available to the public and coupled with a user-friendly portfolio analysis interface for querying the full text and supplemental data. The COVID-19 portfolio is updated daily with new literature selected for inclusion by subject matter experts. It draws upon NLM’s PubMed resource for citations and abstracts of published biomedical literature.
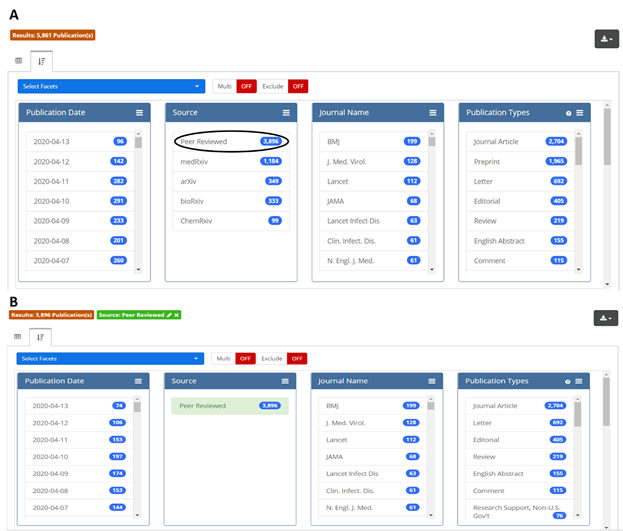
This new resource is designed to provide flexibility and ease-of-use for researchers. Users can take advantage of a full spectrum of Boolean, proximity, and other search methods to query full text and all available supplemental data, or they can limit their search to specific fields, including abstract, author affiliation, or last author. They can also drill down on data once a search is completed. Figure 2 shows a simple example: if a user is interested in analyzing only the peer-reviewed subset of search results, a single click on the “Peer Reviewed” option in the Source facet (circled in black, panel A) will return the desired results (panel B). Some facetable fields, including journal, article type, and author affiliation, are derived from the underlying source data; others, including chemicals & drugs, conditions, and targets, were generated by OPA. All data is downloadable as a CSV or Excel file and includes direct links to the publications and preprints. Also, all searches generate stable URLs that users can share with each other.
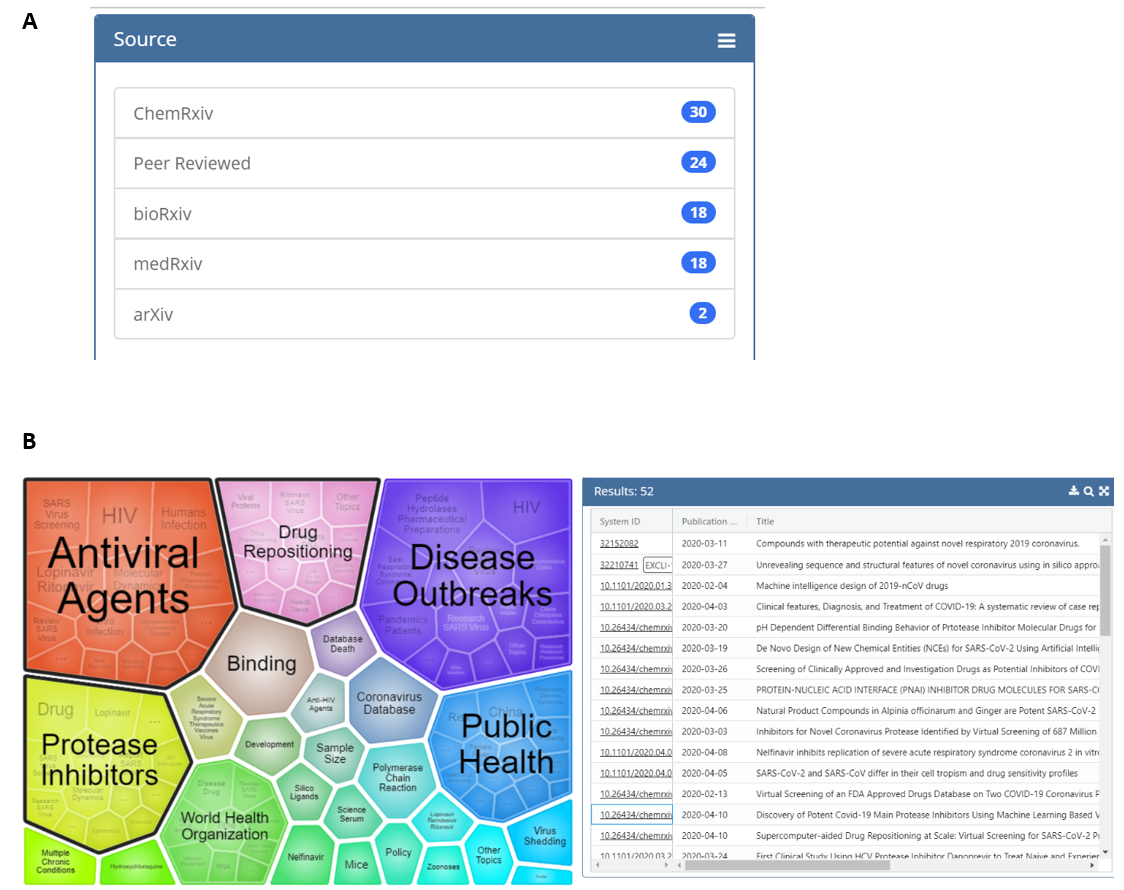
The tool also includes a visualization feature that groups articles into clusters based on key terms. This allows users to obtain, at a glance, the topic areas returned by their search. The clusters in the visualization are also interactive, which allows narrowing of the results to focus on a specific topic of interest. As an example, Figure 3 shows the results of a search of titles, abstracts, full text, and supplemental text with the terms “protease inhibitor” AND ritonavir (search done at 5 pm 4/14/2020); note that a plurality of the results of this search can be found on the ChemRxiv preprint server. Both the visualization image and results can be exported for downstream applications.
We invite you to explore this resource and are excited to see how the research community will use it to gain insight into the COVID-19 outbreak. OPA will continue to add publication sources and features to support the needs of users. Comments are welcome and can be provided directly through the Feedback button at the bottom left of the browser in the tool.
Updated 4/17/20



Great concept and to create some order to the number of reports of dubious credibility and the array of papers of questionable value
I hope there will be a system to prioritize the most relevant articles
look forward to the publication
I look forward to using this as a basis for a seminar topic for my Health Careers Opportunity undergraduate researchers this summer as they conduct online research.
Good but how do we authenticate the results coming? There has to be some coordination.
What is the difference between this and NIH’s LitCovid? https://www.ncbi.nlm.nih.gov/research/coronavirus/
LitCovid is limited to articles in PubMed, includes research on other coronaviruses such as MERS, divides the articles into different categories (e.g. Mechanism, Transmission, Diagnosis, Treatment), and shows the countries of origin on a world map. The iSearch COVID-19 Portfolio tool:
1) includes both publications and preprints (the medRxiv, bioRxiv, ChemRxiv, and arXiv collections; we will soon add the SSRN collection);
2) is curated by subject matter experts to focus coverage on SARS-CoV-2/COVID-19;
3) allows searching of full text and/or supplemental data in addition to titles and abstracts;
4) leverages the cutting-edge analytics available in our iSearch tool, including powerful search functionality and faceting;
5) includes interactive visualizations that allows users to select topics within their search results for download or further queries;
6) makes it easy to download results at any point as a CSV or Excel file.
An example to illustrate points (4) and (5): if you want to find the latest reports about the search for accurate COVID-19 antibody assays, the “minimum should match” search strategy in the iSearch COVID‑19 Portfolio tool is a good approach (this and the other unique analytical methods are explained in the user guide that accompanies the tool). So, entering the following search:
{mm=3} immunoassay antibod* ELISA IgM IgG survey*
currently returns 173 hits; {mm=3} in that query simply means that you’re asking for any records that contain at least three of the terms that follow. The results can be visualized and the “IgM IgG” and “Enzyme-linked immunosorbent assay” subsets transferred to a new window for further exploration of 61 papers, few if any of which are false positives. 5 of those appeared yesterday: 4 preprints (3 medRxiv and 1 bioRxiv) and 1 peer-reviewed PubMed article.
There are positive news that we can get rid of this virus as soon as possible. We are very overwhelmed.
I Dr sushil bargaiya from India.
Similarty between covid19 and pneumocyctic pneumonia.,(pcp)
Pcp is rare in normal immune but very common in weak immune system .2-asymptomatic pcp is extremely common .3-pcp is spread through human to human by air droplet.4-symptomes epidimiolog,chest x-ray.,CT , laboratory finding these all feature are same as covid19 patient।
Pcp is caused by pneumocyctic jiroveci ,this is classified in protozoa kingdom but 1980 when it is seen in hiv patient it is classified in fungus.
I think pneumocyctic jiroveci have both properties of protozoa and fungus .when covid attacks human body it decreases the body immunity and when decrease the immune system this fungus cause pcp – and mortality.
If we use hydroxychloroquine/chloroquine along with cotrimoxazole .we can decreases mortality ….
Consider submitting your idea to the NIH COVID-19 Portal, which collects data on diagnostic, therapeutic, vaccine, and other candidates or technologies with near-term potential for testing against COVID-19, as well as other information that could be leveraged in the response to COVID-19.
Please note that this portal is for information and planning purposes only and shall not be construed as a solicitation or funding opportunity; as a contract, grant, cooperative agreement, or other transaction; or as an obligation on the part of the Federal Government, the NIH, or individual NIH Institutes and Centers to provide support for any ideas identified in response to it. All portions of the submission that are proprietary, confidential, or trade secret should be clearly marked as such.
Thank you so much for consideration of my study
Similarty between covid 19 and pneumocyctic pneumonia.
I pray to God for good health for everyone who are living our planet.
If I will find anything related to covid 19 which can save the people I will share with nih…
Hi,
Good but how do we authenticate the results coming?
There has to be some coordination.
Best Regards.